Welcome to ShinyGO! Just paste your gene list to get enriched GO terms and othe pathways for over 200 plant and animal species. In addition, it also produces KEGG pathway diagrams with your genes highlighted, hierarchical clustering trees and networks summarizing overlapping terms/pathways, protein-protein interaction networks, gene characterristics plots, and enriched promoter motifs. See example outputs below:

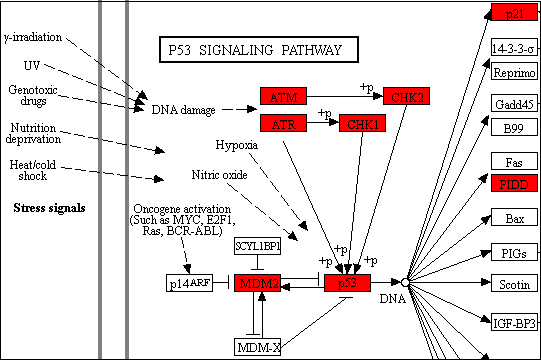

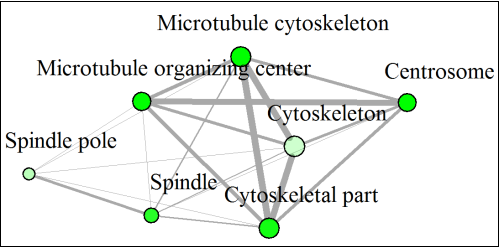

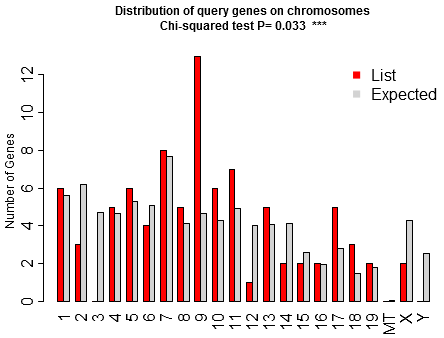

ShinyGO tries to match your species with the 115 archaeal, 1678 bacterial, and 238 eukaryotic species in the STRING server and send the genes. If it is running, please wait until it finishes. This can take 5 minutes, especially for the first time when iDEP downloads large annotation files.
For Details please see our
manuscript
and a detailed
demo.
Based on annotation of 163 animal and 45 plant genomes in Ensembl BioMart as of 12/15/2017.
Additional data from
MSigDB (human),
GSKB (mouse)
and
araPath (arabidopsis).
Full list of supported organisms
For feedbacks or data contributions, please
contact us,
or visit our
homepage.



