Welcome to ShinyGO! Just paste your gene list to get enriched GO terms and othe pathways for over 200 plant and animal species. In addition, it also produces KEGG pathway diagrams with your genes highlighted, hierarchical clustering trees and networks summarizing overlapping terms/pathways, protein-protein interaction networks, gene characterristics plots, and enriched promoter motifs. See example outputs below:



ShinyGO tries to match your species with the 115 archaeal, 1678 bacterial, and 238 eukaryotic species in the STRING server and send the genes. If it is running, please wait until it finishes. This can take 5 minutes, especially for the first time when iDEP downloads large annotation files.
Based on gene onotlogy (GO) annotation and gene ID mapping of
167 animal and 53 plant genomes
in Ensembl BioMart release 93 as of 7/15/2018.
Additional pathway data are collected for some model species from difference sources.
Sources for human pathway databases:
Type
Subtype/Database name
#GeneSets
Source
Gene Ontology
Biological Process (BP)
15796
Ensembl 92
Cellular Component (CC)
1916
Ensembl 92
Molecular Function (MF)
4605
Ensembl 92
KEGG
KEGG
327
Release 86.1
Curated
Biocarta
249
Whichgenes 1.5
GeneSetDB.EHMN
55
GeneSetDB
Panther
168
1.0.4
HumanCyc
240
pathway Commons V9
INOH
576
pathway Commons V9
NetPath
27
pathway Commons V9
PID
223
pathway Commons V9
PSP
327
pathway Commons V9
Recon X
2339
pathway Commons V9
Reactome
2010
V64
Wiki
457
20180610
TF.Target
CircuitsDB.TF
829
V2012
ENCODE
181
V70.0
Marbach2016
628
regulatorycircuits Release 1.0
RegNetwork.TF
1400
7/1/2017
TFacts
428
Feb. 2012
tftargets.ITFP
1926
tftargets May,2017
tftargets.Neph2012
16476
tftargets May,2017
tftargets.TRED
131
tftargets May,2017
TRRUST
793
V2
miRNA.Targets
CircuitsDB.miRNA
140
V. 2012
GeneSetDB.MicroCosm
44
GeneSetDB
miRDB
2588
V 5.0
miRTarBase
2599
V 7.0
RegNetwork.miRNA
618
V. 2015
TargetScan
219
V7.2
MSigDB.Computational
Computational gene sets
858
MSigDB 6.1
MSigDB.Curated
Literature
3465
MSigDB 6.1
MSigDB.Hallmark
hallmark
50
MSigDB 6.1
MSigDB.Immune
Immune system
4872
MSigDB 6.1
MSigDB.Location
Cytogenetic band
326
MSigDB 6.1
MSigDB.Motif
TF and miRNA Motifs
836
MSigDB 6.1
MSigDB.Oncogenic
Oncogenic signatures
189
MSigDB 6.1
PPI
BioGRID
15542
3.4.160
CORUM
2178
02.07.2017
BIND
3807
pathway Commons V9
DIP
2630
pathway Commons V9
HPRD
7141
pathway Commons V9
IntAct
11991
pathway Commons V9
Drug
GeneSetDB.MATADOR
266
GeneSetDB
GeneSetDB.SIDER
473
GeneSetDB
GeneSetDB.STITCH
4616
GeneSetDB
GeneSetDB.T3DB
846
GeneSetDB
SMPDB
699
pathway Commons V9
CTD
8758
pathway Commons V9
Drugbank
2563
pathway Commons V9
Other
GeneSetDB.CancerGenes
23
GeneSetDB
GeneSetDB.MethCancerDB
21
GeneSetDB
GeneSetDB.MethyCancer
54
GeneSetDB
GeneSetDB.MPO
3134
GeneSetDB
HPO
6785
May,2018
Total:
140,438
Sources for mouse pathway databases:
Type
Source
#Sets
Note
Co-expression
Literature
8,742
Differentially expressed genes from 2526
studies
MSigDB
3,964
Molecular Signature Database, v.6.0
L2L
248
List of lists, v.2006.2
CancerGenes*
23
Cancer gene lists
GeneSigDB
494
Gene Signature Database, R.4
Gene
GO_BP
11,943
V2017.5
Ontology
GO_MF
2,932
GO_CC
1,475
Curated
Biocarta*
176
Metabolic and signaling pathways
pathways
PANTHER
151
Ontology-based pathway database, v3.4.1
WikiPathways*
146
Open platform for pathway curation
INOH*
73
Integrating network objects with hierarchies
NetPath*
25
Signal transduction pathways
Metabolic
KEGG
314
Metabolic pathways, R.82.0
pathways
EHMN*
53
Edinburgh human metabolic network
MouseCyc
321
Mouse Biochemical Pathways
, v2013.7
Drug
CTD*
910
The Comparative Toxicogenomics
Database
related
SIDER*
460
Side Effect Resource
MATADOR*
248
Manually Annotated Targets and Drugs Online
Resource
DrugBank*
136
Open data drug and target database
SMPDB*
74
Small Molecule Pathway Database
miRNA
miRDB
1,912
miRNA target prediction and annotations, v
5.0
Target
microRNA.org
314
Predicted miRNA targets, v.R2010
Genes
Grimson et al.
179
Predicted miRNA targets. v.6.2
TarBase
84
Experimentally validated miRNA targets, v.6.0
miRTarBase
775
Experimentally validated miRNA targets, V6.1
MicroCosm
464
Predicted targets
PicTar
35
Predicted miRNA sites, v. 2007.3
TF Target
TFactS*
101
Predicted TF targets
Genes
TRED
99
Confirmed TF target genes, v.2013.7
CircuitsDB
94
Mixed miRNA/TF regulation, v. 2012
TRANSFAC
78
Confirmed TF binding sites, v7.0
Others
Location
341
Genomic location on chromosomes, v.2017
HPO*
1,518
The human phenotype ontology
STITCH*
3,929
Interaction networks of chemicals and
proteins
MPO*
2,943
Mammalian Phenotype Ontology
T3DB*
722
Database of common toxins and their targets
PID*
193
Pathway Interaction Database
MethyCancer*
50
Human DNA methylation and cancer
MethCancerDB*
19
Aberrant DNA methylation in human cancer
Total
46,758
*Secondary data from GeneSetDB
Input:
A list of gene ids, separated by tab, space, comma or new line characters.
Ensembl gene IDs are used internally to identify genes. Other types of IDs will be mapped to Ensembl
gene IDs using ID mapping information available in Ensembl BioMart.
Output:
Enriched GO terms and pathways:

In addition to the enrichment table, a set of plots are produced. If KEGG database is choosen, then enriched pathway diagrams are shown, with user's genes highlighted. Like this one below:

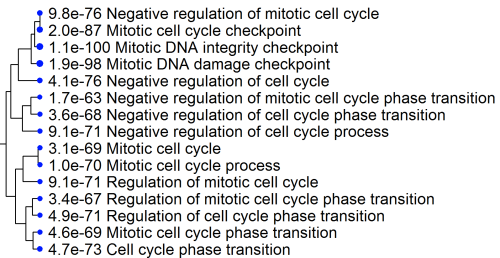
Many GO terms are related. Some are even redundant, like "cell cycle" and "cell cycle process".
To visualize such relatedness in enrichment results, we use a hierarchical clustering tree and network.
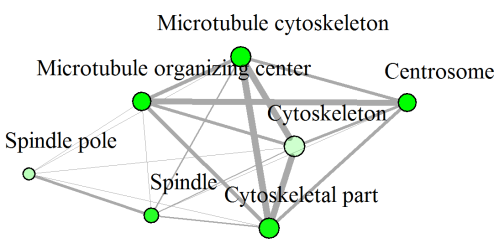
In this tree below, related GO terms are grouped together based on how many genes they share. The size of the solid circle corresponds to the enrichment FDR.

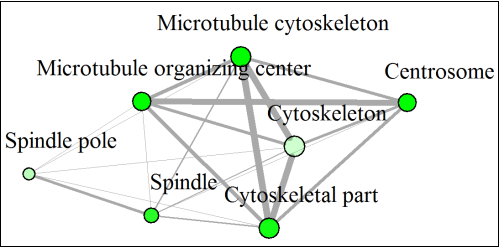
In this network below, each node represent a enriched GO term. Related GO terms are connected by a line, whose thickness reflect percent of overlapping genes. Size of the node corresponds to number of genes.
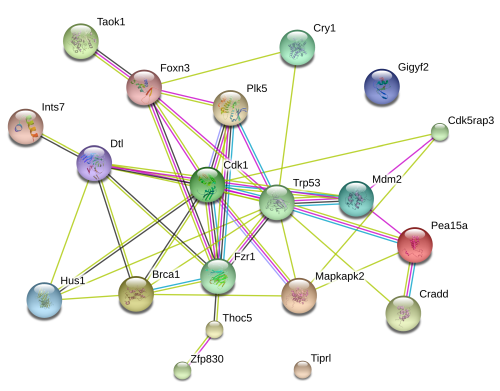
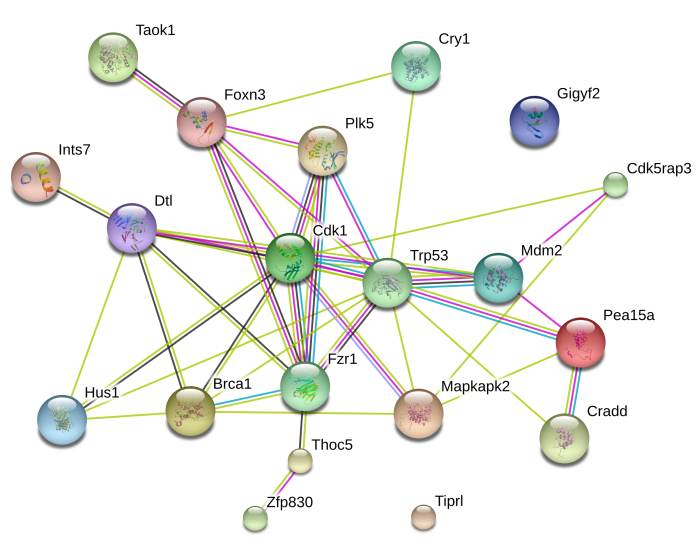
Through API access to STRING-db, we also retrieve protein-protein interaction (PPI) network. In addition to a static network image, users can also get access to an interactive graphics at the www.string-db.org web server.
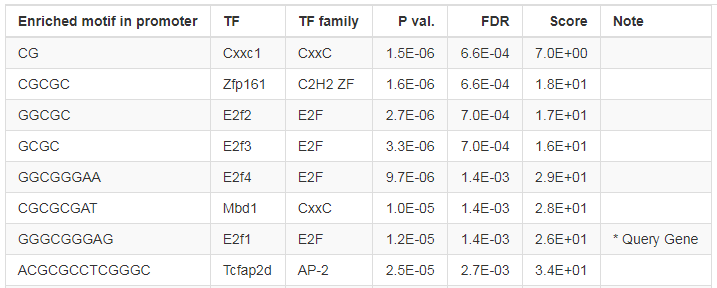
ShinyGO also detects transcription factor (TF) binding motifs enriched in the promoters of user's genes.

Changes:
9/10/2018: V0.5 Upgraded to Ensembl Biomart 92
4/30/2018: V0.42 changed figure configurations for tree.
4/27/2018: V0.41 Change to ggplot2, add grid and gridExtra packages
4/24/2018: V0.4 Add STRING API, KEGG diagram, tree display and network.
|
Type |
Subtype/Database name |
#GeneSets |
Source |
|
Gene Ontology |
Biological Process (BP) |
15796 |
Ensembl 92 |
|
|
Cellular Component (CC) |
1916 |
Ensembl 92 |
|
|
Molecular Function (MF) |
4605 |
Ensembl 92 |
|
KEGG |
KEGG |
327 |
Release 86.1 |
|
Curated |
Biocarta |
249 |
Whichgenes 1.5 |
|
|
GeneSetDB.EHMN |
55 |
GeneSetDB |
|
|
Panther |
168 |
1.0.4 |
|
|
HumanCyc |
240 |
pathway Commons V9 |
|
|
INOH |
576 |
pathway Commons V9 |
|
|
NetPath |
27 |
pathway Commons V9 |
|
|
PID |
223 |
pathway Commons V9 |
|
|
PSP |
327 |
pathway Commons V9 |
|
|
Recon X |
2339 |
pathway Commons V9 |
|
|
Reactome |
2010 |
V64 |
|
|
Wiki |
457 |
20180610 |
|
TF.Target |
CircuitsDB.TF |
829 |
V2012 |
|
|
ENCODE |
181 |
V70.0 |
|
|
Marbach2016 |
628 |
regulatorycircuits Release 1.0 |
|
|
RegNetwork.TF |
1400 |
7/1/2017 |
|
|
TFacts |
428 |
Feb. 2012 |
|
|
tftargets.ITFP |
1926 |
tftargets May,2017 |
|
|
tftargets.Neph2012 |
16476 |
tftargets May,2017 |
|
|
tftargets.TRED |
131 |
tftargets May,2017 |
|
|
TRRUST |
793 |
V2 |
|
miRNA.Targets |
CircuitsDB.miRNA |
140 |
V. 2012 |
|
|
GeneSetDB.MicroCosm |
44 |
GeneSetDB |
|
|
miRDB |
2588 |
V 5.0 |
|
|
miRTarBase |
2599 |
V 7.0 |
|
|
RegNetwork.miRNA |
618 |
V. 2015 |
|
|
TargetScan |
219 |
V7.2 |
|
MSigDB.Computational |
Computational gene sets |
858 |
MSigDB 6.1 |
|
MSigDB.Curated |
Literature |
3465 |
MSigDB 6.1 |
|
MSigDB.Hallmark |
hallmark |
50 |
MSigDB 6.1 |
|
MSigDB.Immune |
Immune system |
4872 |
MSigDB 6.1 |
|
MSigDB.Location |
Cytogenetic band |
326 |
MSigDB 6.1 |
|
MSigDB.Motif |
TF and miRNA Motifs |
836 |
MSigDB 6.1 |
|
MSigDB.Oncogenic |
Oncogenic signatures |
189 |
MSigDB 6.1 |
|
PPI |
BioGRID |
15542 |
3.4.160 |
|
|
CORUM |
2178 |
02.07.2017 |
|
|
BIND |
3807 |
pathway Commons V9 |
|
|
DIP |
2630 |
pathway Commons V9 |
|
|
HPRD |
7141 |
pathway Commons V9 |
|
|
IntAct |
11991 |
pathway Commons V9 |
|
Drug |
GeneSetDB.MATADOR |
266 |
GeneSetDB |
|
|
GeneSetDB.SIDER |
473 |
GeneSetDB |
|
|
GeneSetDB.STITCH |
4616 |
GeneSetDB |
|
|
GeneSetDB.T3DB |
846 |
GeneSetDB |
|
|
SMPDB |
699 |
pathway Commons V9 |
|
|
CTD |
8758 |
pathway Commons V9 |
|
|
Drugbank |
2563 |
pathway Commons V9 |
|
Other |
GeneSetDB.CancerGenes |
23 |
GeneSetDB |
|
|
GeneSetDB.MethCancerDB |
21 |
GeneSetDB |
|
|
GeneSetDB.MethyCancer |
54 |
GeneSetDB |
|
|
GeneSetDB.MPO |
3134 |
GeneSetDB |
|
|
HPO |
6785 |
May,2018 |
|
Total: |
|
140,438 |
|
|
|
|
|
|
|
Type |
Source |
#Sets |
Note |
|
Co-expression |
Literature |
8,742 |
Differentially expressed genes from 2526 studies |
|
|
MSigDB |
3,964 |
Molecular Signature Database, v.6.0 |
|
|
L2L |
248 |
List of lists, v.2006.2 |
|
|
CancerGenes* |
23 |
Cancer gene lists |
|
|
GeneSigDB |
494 |
Gene Signature Database, R.4 |
|
Gene |
GO_BP |
11,943 |
V2017.5 |
|
Ontology |
GO_MF |
2,932 |
|
|
|
GO_CC |
1,475 |
|
|
Curated |
Biocarta* |
176 |
Metabolic and signaling pathways |
|
pathways |
PANTHER |
151 |
Ontology-based pathway database, v3.4.1 |
|
|
WikiPathways* |
146 |
Open platform for pathway curation |
|
|
INOH* |
73 |
Integrating network objects with hierarchies |
|
|
NetPath* |
25 |
Signal transduction pathways |
|
Metabolic |
KEGG |
314 |
Metabolic pathways, R.82.0 |
|
pathways |
EHMN* |
53 |
Edinburgh human metabolic network |
|
|
MouseCyc |
321 |
Mouse Biochemical Pathways , v2013.7 |
|
Drug |
CTD* |
910 |
The Comparative Toxicogenomics Database |
|
related |
SIDER* |
460 |
Side Effect Resource |
|
|
MATADOR* |
248 |
Manually Annotated Targets and Drugs Online Resource |
|
|
DrugBank* |
136 |
Open data drug and target database |
|
|
SMPDB* |
74 |
Small Molecule Pathway Database |
|
miRNA |
miRDB |
1,912 |
miRNA target prediction and annotations, v 5.0 |
|
Target |
microRNA.org |
314 |
Predicted miRNA targets, v.R2010 |
|
Genes |
Grimson et al. |
179 |
Predicted miRNA targets. v.6.2 |
|
|
TarBase |
84 |
Experimentally validated miRNA targets, v.6.0 |
|
|
miRTarBase |
775 |
Experimentally validated miRNA targets, V6.1 |
|
|
MicroCosm |
464 |
Predicted targets |
|
|
PicTar |
35 |
Predicted miRNA sites, v. 2007.3 |
|
TF Target |
TFactS* |
101 |
Predicted TF targets |
|
Genes |
TRED |
99 |
Confirmed TF target genes, v.2013.7 |
|
|
CircuitsDB |
94 |
Mixed miRNA/TF regulation, v. 2012 |
|
|
TRANSFAC |
78 |
Confirmed TF binding sites, v7.0 |
|
Others |
Location |
341 |
Genomic location on chromosomes, v.2017 |
|
|
HPO* |
1,518 |
The human phenotype ontology |
|
|
STITCH* |
3,929 |
Interaction networks of chemicals and proteins |
|
|
MPO* |
2,943 |
Mammalian Phenotype Ontology |
|
|
T3DB* |
722 |
Database of common toxins and their targets |
|
|
PID* |
193 |
Pathway Interaction Database |
|
|
MethyCancer* |
50 |
Human DNA methylation and cancer |
|
|
MethCancerDB* |
19 |
Aberrant DNA methylation in human cancer |
|
|
Total |
46,758 |
*Secondary data from GeneSetDB |