Figure width can be adjusted by changing the width of browser window.
Figure width can be adjusted by changing the width of browser window.
Figure width can be adjusted by changing the width of browser window.
Figure width can be adjusted by changing the width of browser window.
QC reports are not available for custom organisms.
Genes types are based on ENSEMBL classification.
Higher proportions of rRNA indicuate ineffective rRNA-removal.
Higher bar means more reads map to this chromosome in this sample.
A tall bar means genes on this chromosome are expressed at higher levels in a sample, as indicated by the 75th percentile.
Figure width can be adjusted by changing the width of browser window.
In the pre-process stage, we (1) filter out genes with extremely low expression, (2) Gene IDs are converted to Ensembl gene ID or STRING-db gene ID, and (3) apply transformations.
Counts data
Filtering: Some genes are not expressed in any samples. Others are expressed at extremely low levels. We need to remove these genes from further analysis. By default, a gene must have more than 0.5 counts per million (CPM) in at least one sample. Otherwise, the gene is removed. Users can specify that genes must be above 0.5 CPM in two samples by changing the "n libraries" option to 2.
CPMs are calculated by normalizing the read counts by the total counts per sample. For example, if sample 1 has 20 million counts in total, then the read counts are divided by 20 to get the CPM.
The R command used for filtering is this.
x <- x[ which( apply( cpm(DGEList(counts = x)), 1, function(y) sum(y>=minCPM)) >= nLibraries ) , ]
The data is normalized by cpm function in edgeR. The number of samples above a minCPM is counted. Only genes with levels above minCPM in at least nLibraries are retained.
It is very common to filter out 30% or even 50% of the genes on the genome-wide expression profiles, as we do not expect all genes in the genome to be expressed in one particular tissue/cell. Keeping lowly expressed genes just adds noise to downstream analyses. But aggressive filtering may lead to missing some genes that are expressed low but are significantly different. minCPM can be reduced if your library size is big. For example, if your library size is 50 million on average, you can lower your minCPM to 0.2. This will eliminate genes represented at least by 10 reads.
Transformation: There are 3 options for the transformation of counts data for clustering analysis and principal component analysis:
- VST: variance stabilizing transform (Anders & Huber 2010)
- rlog: regularized log (only for N<10, see Love et al. 2014)
- Started log: log2(x+c)
VST is performed according to (Anders and Huber 2010) and rlog is based on (Love, Huber et al. 2014). When there are more than 10 samples, rlog becomes slow. The default is started log, where a pseudo count c is added to all counts before log transformation. The constant c can range from 1 to 10. The bigger this number is, the less sensitive it is to noise from low counts. We have found c=4 offer balanced transformation based on several datasets we tried. The started log transformation is equivalent to the logCPM offered in edgeR. The only difference is that the libraries are not all scaled to 1 million per sample. The libraries are scaled first using DESeq2's normalized = TRUE option:
x <- log2( counts(dds, normalized=TRUE) + c )
The effect of these transformations can be visualized below between technical replicates. You can see that VST is very aggressive in transforming the data. The rlog option is slower, especially when there are many samples, but it makes the distribution across samples closer than a simple log.
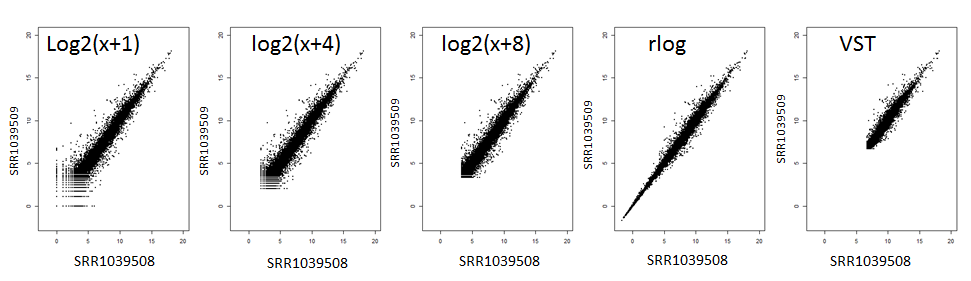
For counts data, transformed data is used for exploratory data analysis (EDA, such as clustering analysis and PCA, MDS). If you choose limma-trend to identify differentially expressed genes (DEGs), then transformed data is also used. Both DESeq2 and limma-voom use original read counts data, not the transformed data.
iDEP produces a barplot representing total read counts per library. When the library sizes are more than 3 times different, limma-trend method is not recommended for identifying DEGs. See the manual for limma .

Bias in sequencing depth can exist in samples. The above is an example, where fewer reads in the p53 wild-type samples after radiation treatment. An ANOVA is conducted by iDEP and if P<0.01, a warning is produced. This presents a confounding factor. iDEP also calculates the ratio of the max/min total counts among groups. It is believed that as long as the ratio is within 1.5, it should be acceptable. See this discussion on Twitter .
FPKM, microarray or other normalized expression data
For normalized expression data, a filter is also applied to remove genes expressed at low levels across all samples. For example, users can choose to include only genes expressed at the level of 1 or higher in at least one sample. This number works for FPKM or RPKM data, but it should be changed according to the data format. For cDNA microarrays, where the expression levels are log ratios, we need to set this to a negative number such as -1e20 to disable this filter.
This filter is very important as it affects downstream analyses. For FPKM, RPKM data, many people choose 1 as a cutoff. For genome-wide expression analysis using DNA microarray, it is safe to choose a cutoff that removes the bottom 20-40% of genes when ranked by maximum expression level across samples in descending order.
iDEP calculates kurtosis for each of the data columns, and if the mean kurtosis is bigger than 50, a log2 transformation is enforced. Large kurtosis usually indicates the presence of extremely large numbers in the data set that warrants log transformation.
Users can double-check the effects of data transformation by examining the box plot and density plot on this page.
References:
Anders, S. and W. Huber (2010). "Differential expression analysis for sequence count data." Genome Biol 11(10): R106.
Love, M. I., W. Huber and S. Anders (2014). "Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2." Genome Biol 15(12): 550.
Broaden your browser window if there is overlap -->
Cutoff:
Two pathways (nodes) are connected if they share 30% (default, adjustable) or more genes. Green and red represents up- and down-regulated pathways. You can move the nodes by dragging them, zoom in and out by scrolling, and shift the entire network by click on an empty point and drag. Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes.
Note: In the gene type column, "C" indicates protein-coding genes, and "P" means pseduogenes.
Details of Enrichment Analysis
Using genes with maximum expression level at the top 75%. Data is transformed and clustered as specified in the sidebar.
Numbers of differentially expressed genes for all comparisons. "B-A" means B vs. A. Interaction terms start with "I:"
The above graph is an UpSet plot that is an alternative to a venn diagram. The plot shows the intersections of the data in the combination matrix (bottom) and the columns show how many genes are in each intersection.
More info on plot
Cutoff:
Two pathways (nodes) are connected if they share 30% (default, adjustable) or more genes. Green and red represents up- and down-regulated pathways. You can move the nodes by dragging them, zoom in and out by scrolling, and shift the entire network by click on an empty point and drag. Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes.
Note: In the gene type column, "C" indicates protein-coding genes, and "P" means pseduogenes.
Details of Enrichment Analysis
Adjusting the width of the browser window can render figure differently and resolve the "Figure margin too wide" error.
Connected gene sets share more genes. Color of node correspond to adjuested Pvalues.
Cutoff:
Two pathways (nodes) are connected if they share 30% (default, adjustable) or more genes. Green and red represents up- and down-regulated pathways, respectively. You can move the nodes by dragging them, zoom in and out by scrolling, and shift the entire network by click on an empty point and drag. Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes.
Please select KEGG database, if available, from left and perform pathway analysis first.
Introduction
Pathway analyses are done using fold-change values of all genes calculated by limma or DESeq2. This is different from the enrichment analysis in the DEG2 tab, which only uses gene lists of differentially expressed genes (DEGs).
Our method also differs from traditional methods like Gene Set Enrichment Analysis (GSEA), which takes the normalized expression matrix and conducts more sophisticated analyses like permutation tests. For confirmation, please use the stand-alone GSEA.
Methods
Pathway analysis can be performed using several methods. GSEA (Gene Set Enrichment Analysis) (Subramanian et al., 2005) is conducted in the pre-ranked mode using a recent faster algorithm based on the fgsea package (Sergushichev, 2016). PAGE (Parametric Analysis of Gene Set Enrichment) (Kim and Volsky, 2005) is used as implemented in PGSEA package (Furge and Dykema, 2012). For PGSEA there are two versions. One only analyzes the selected comparisons and another option (“PGSEA w/ all samples”) enables the user to analyze all sample groups.
Unlike all these methods that rely on the built-in geneset databases, ReactomePA (Reactome Pathway Analysis) (Yu and He, 2016) retrieves genesets from Reactome (Fabregat et al., 2016; Yu and He, 2016).
Gene expression data can be visualized on KEGG pathway diagrams (Kanehisa et al., 2017) using Pathview (Luo and Brouwer, 2013). Note that Pathview download pathway diagrams directly from KEGG website and thus is slow.
On the lower left side of the screen, there is a checkbox named “Use absolute values of fold changes for GSEA and GAGE”. This is useful as some molecular pathways can be modulated by up-regulating some genes while down-regulating others. This is especially useful when using KEGG pathways. For other genes sets such as TF target genes and microRNA target genes where we know the regulation is one-directional, we should not check this box.
Users can choose to filter out some genes with noisy fold-changes by using an FDR cutoff by reducing the “Remove genes with big FDR before pathway analysis” from the default value of 1 to a relatively bigger FDR cutoff like 0.6. If this is done, genes with FDR > 0.6 is removed from pathway analysis.
Interpretation
iDEP makes it easy for users to choose different pathway analysis methods and databases. The drawback is that it is easy to rationalize any a priori hypothesis (bias). With so many choices and knobs to turn, you can make any story you wanted to make!
Results from pathway analysis do not directly inform about the activity of the pathway of interest but often reflect downstream effects of changes in the activities of transcription factors.
Pathway Database
Very small genesets can cause problems. By default, genesets with less than 15 genes are disregarded. Sometimes this needs to be reduced to 10, or even 5, when some pathways of interest only has few genes. But be aware that this may introduce false positives.
Besides Gene Ontology, iDEP includes additional data from KEGG, Reactome, and many other sources. There might be errors in our conversion of these diverse databases.
R Code
R code used for GAGE using the gage package:
paths <- gage(fold, gsets = gmt, ref = NULL, samp = NULL)
R code for GSEA via fgsea pckage:
paths <- fgsea(pathways = gmt,
stats = fold,
minSize=input$minSetSize,
maxSize=input$maxSetSize,
nperm=5000)
R code for PAGE using the PGSEA package:
pg= PGSEA (convertedData – rowMeans(convertedData), cl=gmt, range=myrange, p.value=TRUE, weighted=FALSE)
R code for ReactomePA:
paths <- gsePathway(fold, nPerm=50000, organism = ReactomePASpecies[ix],
minGSSize= input$minSetSize,
maxGSSize= input$maxSetSize,
pvalueCutoff=0.5,
pAdjustMethod=”BH”, verbose=FALSE)
References
Fabregat, A., Sidiropoulos, K., Garapati, P., Gillespie, M., Hausmann, K., Haw, R., Jassal, B., Jupe, S., Korninger, F., McKay, S., et al. (2016). The Reactome pathway Knowledgebase. Nucleic Acids Res 44, D481-487.
Furge, K., and Dykema, K. (2012). PGSEA: Parametric Gene Set Enrichment Analysis. R package version 1480.
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353-D361.
Kim, S.Y., and Volsky, D.J. (2005). PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics 6, 144.
Luo, W., and Brouwer, C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830-1831.
Sergushichev, A. (2016). An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv http://biorxiv.org/content/early/2016/06/20/060012.
Subramanian, A., Tamayo, P., Mootha, V.K., Mukherjee, S., Ebert, B.L., Gillette, M.A., Paulovich, A., Pomeroy, S.L., Golub, T.R., Lander, E.S., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545-15550.
Yu, G., and He, Q.Y. (2016). ReactomePA: an R/Bioconductor package for Reactome pathway analysis and visualization. Mol Biosyst 12, 477-479.
Select a region to zoom in. Mouse over the points to see more information on the gene. Enriched regions are highlighted by blue or red line segments paralell to the chromosomes.
Mouse over the figure to see more options on the top right, including download.
Where are your differentially expressed genes (DEGs) located on the genome?
Red and blue dots represent significantly up- and down-regulated genes, respectively, according to the criteria on the side panel. These criteria could differ from the one in DEG1 tab. The distance of the dots from the closest chromosome is proportional to the log2 fold-change (FC).
Are there regions of the genome where genes are coherently up- or down-regulated?
To answer this question, we scan the genome with sliding windows. Within each window we take several steps to slide forward. For example if you choose a window size = 6Mbps and steps = 3, the first window is from 0 to 6 Mbps, the 2nd from 2 to 8Mbps, and the third from 4 to 10 Mbps, and so on.
For all genes in a window/region, we test whether the mean of FC of these genes is zero using a t-test. All genes analyzed by DESeq2 or limma, significant or otherwise, are included in this analysis. Hence this result is indepdent of our DEG cutoffs. P values from the test of the mean are adjusted to FDR. Essentially, we considered genes located in a genomic region as a gene set or pathway, and we performed simple pathway analysis by asking whether these genes are behaving non-randomly.
Based on an FDR cutoff for the windows, red and blue segments indicate genomic regions with genes coherently up- or down-regulated, respectively. Below you can adjust the window size, and steps in a window, and FDR cutoff for windows. Mouse over to see gene symbols or IDs. Zoom in regions of interest. The chromosomes may be only partly shown as we use the last gene's location to draw the line.
Enriched pathways in the selected cluster:
Cutoff:
Two pathways (nodes) are connected if they share 30% (default, adjustable) or more genes. Green and red represents up- and down-regulated pathways. You can move the nodes by dragging them, zoom in and out by scrolling, and shift the entire network by click on an empty point and drag. Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes.
Note: In the gene type column, "C" indicates protein-coding genes, and "P" means pseduogenes.
Details of Enrichment Analysis
Enriched pathways in the selected module
Cutoff:
Two pathways (nodes) are connected if they share 30% (default, adjustable) or more genes. Green and red represents up- and down-regulated pathways. You can move the nodes by dragging them, zoom in and out by scrolling, and shift the entire network by click on an empty point and drag. Darker nodes are more significantly enriched gene sets. Bigger nodes represent larger gene sets. Thicker edges represent more overlapped genes.
Note: In the gene type column, "C" indicates protein-coding genes, and "P" means pseduogenes.
Details of Enrichment Analysis
If you find iDEP helpful, please send us a brief email (gelabinfo@gmail.com).
If you state your general research area and how iDEP makes you more productive, we can use it as a support letter when we apply for the next round of funding. Hundreds of strong, enthusiastic letters sent to us in 2019 were essential when we applied for the current grant from NIH/NHGRI (R01HG010805), which expires in 20 months. Your letters will help sustain and improve this service.
iDEP is developed and maintained by a small team at South Dakota State University (SDSU). Our team consists of Xijin Ge (PI), Jianli Qi (research staff), and several talented students. None of us are trained as software engineers. But we share the passion about developing an user-friendly tool for all biologists, especially those who do not have access to bioinformaticians.
Graduate students contributed to this project include Eun Wo Son, Runan Yao, Roberto Villegas-Diaz, Eric Tulowetzke, Emma Spors, Chris Trettel, and Ben Derenge. Undergraduate students include Jenna Thorstenson and Jakob Fossen. Research staff include Jianli Qi and Gavin Doering. Much of the new version of iDEP is rewritten by Gavin Doering. The iDEP logo was designed by Emma Spors. Technical support is kindly provided by the Office of Information Technology (OIT) at SDSU. Mirror site is enabled by a JetStream2 allocation award (BIO210175), which is supported by NSF.
Cite the iDEP paper, otherwise, this service might vanish!
If you use iDEP, even just for prelimiary analysis, please cite: Ge, Son & Yao, iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data, BMC Bioinformatics 19:1-24, 2018. Merely mentioning iDEP with an URL is insufficient. It is difficult to track. Consider citing other tools that form the foundation of iDEP, such as ENSEMBL, STRING-db, DESeq2, limma and many others. If you use the KEGG diagram, please also cite pathview, and KEGG.
According to Google Scholar, more than 900 papers cited iDEP, as of April 19, 2024. Our website has been accessed over 600,000 times by 120,000 users, spending 15 minutes each time. For every 1000 users, only 6 cited the iDEP paper, which is disappointingly low.
Source code, database, & local installation
Source code is available on GitHub, which also includes instructions to install iDEP on your local machine using our database.
Reproducibility
Download the reports in the multiple tabs, which has the parameters and results. Also download and try the the R code on the DEG1 tab.
Previous versions of iDEP are still useable:
iDEP 1.13 with Ensembl Release 107, archived January 5, 2024iDEP 0.96 with Ensembl Release 104, released on July 30, 2022
iDEP 0.95 with Ensembl Release 104, released on Feb. 8, 2022
iDEP 0.94 with Ensembl Release 104, released on Oct. 15, 2021
iDEP 0.93 with Ensembl Release 103, released on May 20, 2021
iDEP 0.92 with Ensembl Release 100, released on May 20, 2021
iDEP 0.90 with Ensembl Release 96, released on May 19, 2021
iDEP 0.85 with Ensembl Release 95, released on March 29, 2019
iDEP 0.82 with Ensembl Release 92, released on July 11, 2018
iDEP 0.73 with Ensembl Release 91, released in December 2017
Privacy policy
User uploaded data files are saved in a temporary folder during your session and automatically deleted. Our group does not keep a copy of the uploaded data. We monitor web traffic using Google Analytics, which tells us your IP address (approximate location down to the city level), and how long you are on this site. Error messages are recorded by Shiny server. By visiting this site, you agree to provide web activity data.
Contact us
Please email Jenny gelabinfo@gmail.com (recommended) or Dr. Ge xijin.ge@sdstate.edu (unreliable). Follow us on Twitter for recent updates. File bug reports on GitHub.
NO GUARANTEE OF ACCURACY
iDEP is developed by a small team with limited resources. We have not thoroughly tested it. So please verify all findings using other tools or R scripts. We tried our best to ensure our analysis is correct, but there is no guarantee.
By offering so many combinations of methods to analyze a data set, iDEP enables users to rationalize. It is human nature to focus on results that we like to see (confirmation bias). It is unfortunate that you can almost found further support for almost any theory from the massive but noisy literature. We encourage users to be critical of the results obtained using iDEP. Try to focus on robust results, rather than those that only should up with a certain parameter using a particular method.
Change log
4/20/2024: Fix bug in network tab related to module download. Enable download of network as a CSV file.
4/19/2024: iDEP 2.01. Minor upgrade. Fixed a bug related to insufficiant # of color in palettes. Optimized UI for load data. Reverted to basic Shiny theme due to an issue with new version of Shiny package.